Reveal the epigenomic landscape, cell by cell
Chromium Single Cell ATAC (Assay for Transposase Accessible Chromatin) allows you to analyze chromatin accessibility at the single cell level, providing insights into cell types and states, and deeper understanding of gene regulatory mechanisms.

Workflow
1. Prepare your sample
Start with a nuclei suspension isolated from cell culture, primary cells, or fresh or frozen tissue.
2. Construct Your 10x Library
Construct a 10x barcoded library using our reagent kits and a compatible Chromium instrument. Each member of the Chromium instrument family encapsulates each cell with a 10x barcoded Gel Bead in a single partition. Within each nanoliter-scale partition, cells undergo reverse transcription to generate cDNA, which shares a 10x Barcode with all cDNA from its individual cell of origin.
3. Sequence
The resulting 10x Barcoded single cell ATAC-seq library is compatible with standard NGS short-read sequencing on Illumina sequencers, for massively parallel epigenomic profiling of thousands of individual cells.
4. Analyze Your Data
Our Cell Ranger ATAC analysis software generates open chromatin profiles for each cell, identifies clusters of cells with similar profiles and calls peaks, and can aggregate data from multiple samples.
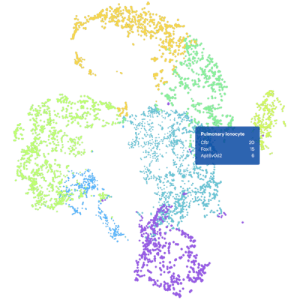
5. Visualize Your Data
Use our Loupe visualization software to interactively explore your results, perform differential accessibility analysis, find clusters enriched for motifs of interest, and compare across samples.